- Published at
Weingarten Map
This post provides theoretical foundation behind the Weingarten map for curvature estimation along with its Python implementation for 2D images.
- Authors

- Name
- Dr. Dmitrij Sitenko
- Mathematical Image Processing at Ruprecht Karls University Heidelberg
Table of Contents
The measure of curvature at a point on a surface is an intrinsic property—it depends only on the distances on the surface and is independent of how the surface is embedded in space.
Carl Friedrich Gauss
Let be an -dimensional submanifold in a -dimensional Euclidean space with an induced Riemannian metric. At each point , consider the decomposition of the tangent space:
For each normal vector , let be the extension of to the whole manifold . The Weingarten map or shape operator at with respect to is the mapping:
where is the orthogonal projection to the tangent space. An important property of the Weingarten map is that it is independent of the particular choice of extension .
Expanding the above map using a Taylor series yields:
Theorem
This makes it an attractive feature descriptor for measuring the variation of the normal vector field of a given dataset. We next illustrate how this is performed on an image data set and provide a guidelilne for an easy implementation in python.
Given an image , we first estimate the basis of the tangent and normal spaces at pixel by constructing the covariance matrix
from which we can estimate a smooth normal vector field for each pixel through through eigenvalue decomposition.
import numpy as np
from scipy.ndimage.filters import convolve
def Weingarten_Map(I,Mask_dim_x = 11,Mask_dim_y = 11):
assert Mask_dim_x % 2 != 0 and Mask_dim_y % 2 != 0
Mask_dim_x = 11
Mask_dim_y = 11
N = (Mask_dim_x*Mask_dim_y)
Mask = np.ones((Mask_dim_x,Mask_dim_y))/N
Res_x = 1/I.shape[0]
Res_y = 1/I.shape[1]
Im_pred_vec = np.zeros((*I.shape,3))
Im_pred_vec_mean = np.zeros((*I.shape,3))
Im_pred_vec[:,:,2] = I[:,:]
Im_pred_vec[:,:,0:2] = np.moveaxis(np.array(np.indices((Im_pred_vec.shape[0:2])),dtype = np.float32),0,-1)
' Add Resolution to the indices '
Im_pred_vec[:,:,0:2] = np.einsum("ijk,k->ijk",Im_pred_vec[:,:,0:2],np.array([Res_x,Res_y]))
Im_pred_vec_mean = convolve(Im_pred_vec,Mask[:,:,None])
' Compute Covariance Matrix '
Im_pred_vec[:,:,0:2] = np.einsum("ijk,k->ijk",Im_pred_vec[:,:,0:2],np.array([Res_x,Res_y]))
Im_pred_vec_mean = convolve(Im_pred_vec,Mask[:,:,None])
Im_pred_vec_mean_w = convolve(Im_pred_vec,Mask[:,:,None])
Cov_Mat = np.zeros((*I.shape,3,3))
Cov_Mat = N*convolve(np.einsum('ijk,ijl->ijkl', Im_pred_vec, Im_pred_vec), Mask[:, :, None,None], mode="nearest")
Cov_Mat -= np.einsum('ijk,ijl->ijkl', Im_pred_vec_mean_w, Im_pred_vec_mean)
Cov_Mat -= np.einsum('ijl,ijk->ijkl', Im_pred_vec_mean_w, Im_pred_vec_mean)
Cov_Mat += np.einsum('ijk,ijl->ijkl', Im_pred_vec_mean, Im_pred_vec_mean)
The first largest eigenvectors of covariance matrix determine a basis for the tangent space approximation for each pixel location . Hereby, the last eigenvectors determine a basis of corresponding normal space. Lets reproduce this part
'Compute normals and tangets for each pixel location'
N_space,T_space,_ = np.split(np.linalg.eigh(Cov_Mat)[1],(1,3),axis = 3)
N_space = np.squeeze(N_space)
To compute the Weingarten map, we need to extend the normal vector field smoothly across entire submanifold . This is achieved by projecting to normal spaces at each pixel which results in a smooth normal vector field
Consider a twice-differentiable function supported on the interval and a parameter which defines a scaled kernel:
Leveraging the tangent basis at pixel and assuming that is close to , the Result simplifies to the following first-order approximation of the Weingarten map:
where . As a consequence, in order to get access to the Weingarten map need to minimize the objective
Introducing the auxiliary mappings:
solution to optimization problem (1) exhibits a closed-form solution such that the Weingarten map at each pixel is represented in terms of the decomposition
Note, that we need to compute for all pixels which involves the sum over all pixel to evaluate the objective (1).
This results in large dimension of matrices in (2). Due to the exponential decay of the kernel function we instead use an approximation by summing over a suffieciently large neighbourhood.
N_space_pad = np.pad(N_space,((X_shift-1,X_shift-1),(Y_shift-1,Y_shift-1),(0,0)))
N_space_pad = view_as_windows(N_space_pad,(Mask_dim_x,Mask_dim_y,3),step=(1, 1, 1))
N_space_pad = np.squeeze(N_space_pad)
Delta_k = np.einsum('ijlk,ijsml->ijsmk',T_space[:,:,:,:],X_diff)
Scalar_Mat = np.einsum('ijsmk,ijk->ijsm',N_space_pad,N_space)
N_space_pad = N_space_pad*Scalar_Mat[:,:,:,:,None]-N_space[:,:,None,None,:]
Theta_k = np.einsum('ijkl,ijsmk->ijsml',T_space,N_space_pad)
Diag_W = np.exp(-np.einsum('ijkls,ijkls->ijkl',X_diff,X_diff)/(sigma**2))/(sigma**2)
W_Matrix = -np.einsum('ijsmk,ijsml->ijkl',Theta_k*Diag_W[:,:,:,:,None],Delta_k)
Theta_k = np.einsum('ijsmk,ijsml->ijkl',Delta_k*Diag_W[:,:,:,:,None],Delta_k)+0.1*np.eye(2)[None,None,:,:]
Theta_k = np.linalg.inv(Theta_k.reshape(-1,2,2)).reshape(*Theta_k.shape)
W_Matrix = np.einsum('ijsk,ijkm->ijsm',W_Matrix,Theta_k)
W_Matrix = 0.5*(W_Matrix+np.swapaxes(W_Matrix,axis1=2,axis2=3))
Given the Weingarten map, we gain access to the second fundamental form, mean curvature, and sectional curvature at each image location . These features can be used for classification and segmentation tasks.
Gauss Formuala and the Second Fundamental Form
For each submanifold with the corresponding induced Riemannian metric we denote by the connection on and the connection on . Then for any pair of vector fields we have the Gauss formula.
the map is a second-order symmetric covariant tensor field on commonly known as the second fundamental form of submanifold . For each normal vector field the second fundamental form and the Weingarten are related by the Weingarten formula
with
The following plot illustrates the computed curvature measurues that can be used for a feature extraction filter bank
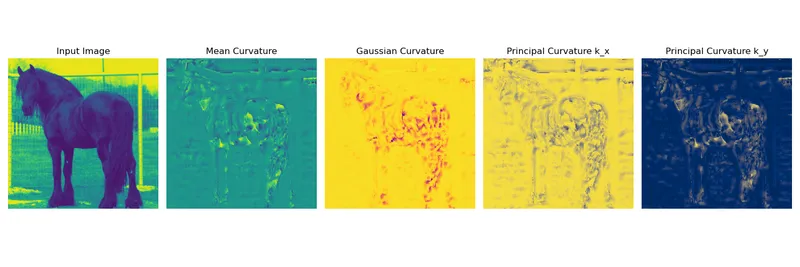
Curvature
The Weingarten Map is a symmetric matrix that encodes the curvature information of a surface. Using , we can define the following curvatures:
Principal Curvatures
The principal curvatures and are the eigenvalues of , given by:
where:
- is the trace of ().
- is the determinant of ().
Mean Curvature
The mean curvature is the average of the principal curvatures:
Gaussian Curvature
The Gaussian curvature is the product of the principal curvatures:
nx,ny = W_Matrix.shape[0:2]
mean_curvature = np.zeros((nx, ny))
gaussian_curvature = np.zeros((nx, ny))
principal_curvatures = np.zeros((nx, ny, 2)) # Store kappa1, kappa2
# Compute eigenvalues and curvatures
for i in range(nx):
for j in range(ny):
W = W_Matrix[i, j]
# Compute eigenvalues (principal curvatures)
kappa1, kappa2 = np.linalg.eigvalsh(W) # Use eigvalsh for symmetric matrices
principal_curvatures[i, j] = [kappa1, kappa2]
# Mean and Gaussian curvatures
mean_curvature[i, j] = 0.5*(kappa1 + kappa2)
gaussian_curvature[i, j] = kappa1 * kappa2